How ordering works
To see prices and place an order for our Next-Generation Sequencing services (including library prep), please use our Laboratory Information Management System (LIMS), SeqLIMS. At any stage in SeqLIMS, you can hover over the help icon for more details. For questions about this process, please contact us.
If you have any project design or technical questions, you can contact us by email or schedule a consultation.
Create an Account in SeqLIMS

- Go to SeqLIMS and click on "Sign up."
- Enter name, email for username, and password.
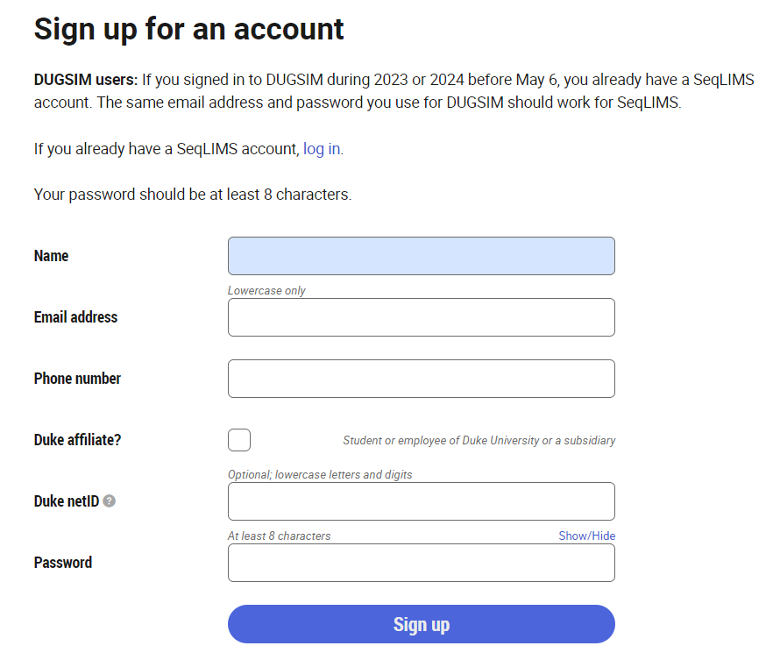
*Checkmark Duke Affiliate? Enter your Duke NetID, if you have one. Otherwise, leave the textbox empty.
Get an Official Quote (Account Required)
- Log into SeqLims

- Click on "New Project"

- Click the drop-down menu for "Service" and choose sequencing platform
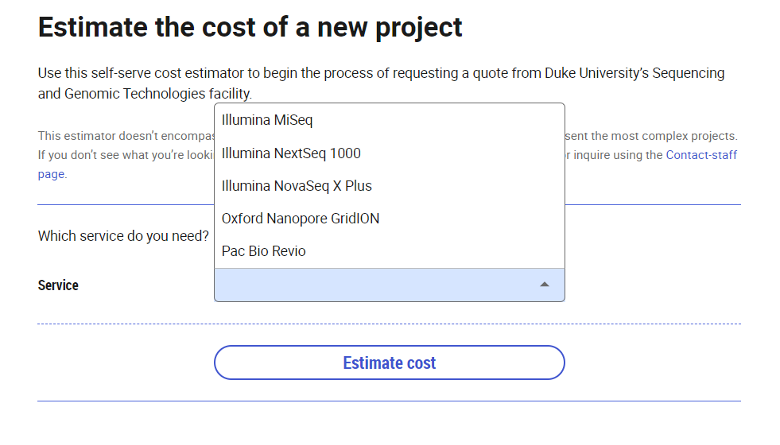
- Select the type of sample for submission. Specimens are cell pellets, blood, and nucleic acids.

- For each sample type, input the sample quantity (number of samples), sequencing product (instrument and flow cell or lane), and sequencing product quantity (number of flow cells or lanes). If you require nucleic acid extractions, select specimen-processing product (e.g. RNA extraction from cell pellet). If you require library-preparation, select library-preparation product for the type of library build (e.g. mRNA-seq).
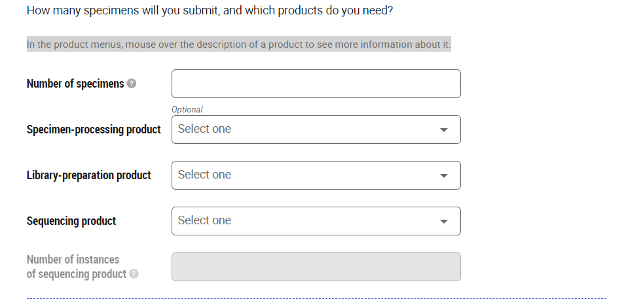
- Click on the "Estimate quote" button at the bottom of the page.

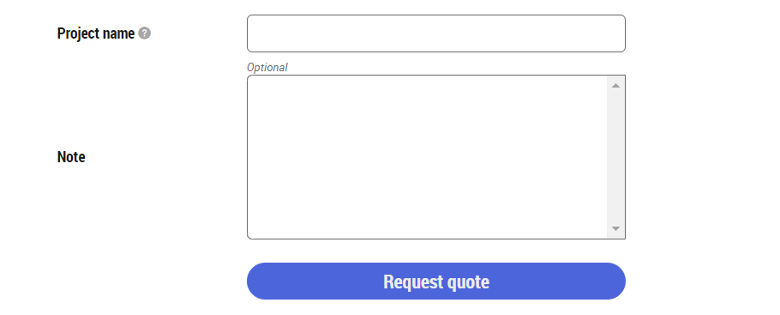
- Review the estimate cost. Enter a project name, additional notes, questions, or information, if any, and click on the "Request quote" button at the bottom of the page.
- Our staff will review and process your request within 24 hours.
Place an Order (Quote Required)
- You will receive an email from SeqLIMSof your approved quote.
- Sign into SeqLIMS.
- Go to the "Main" page and locate your quote.
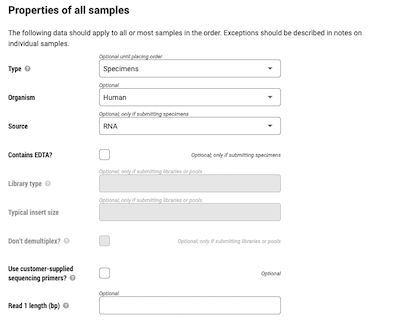
- Input properties of samples. To help ensure the best experiment, provide as much detail as possible.
- For client prepared libraries, click the button acknowledging that sequencing results for client-built libraries are not guaranteed. This button is required to place an order

- Scroll to the bottom of the page and directly place an order from the approved quote by clicking "Convert quote into order."

*Non-Duke customers must also include information of an authorized representative of your institution, that is, a person able to bind the institution to the agreement. 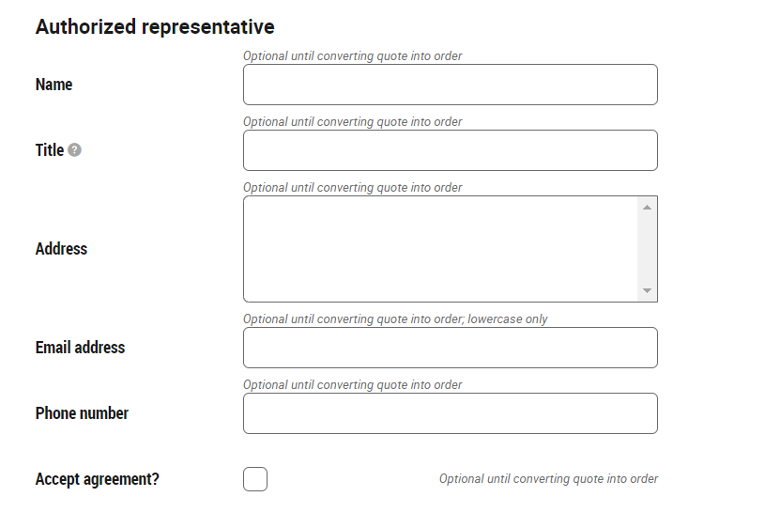
- Once an order is placed and we have validated the mode of payment (reception of your Purchase Order or valid Fund Code), a project will be assigned a unique order number. We do not accept credit cards.
Prepare Your Sample(s) (Order Required)
*We will return samples that are not properly prepared and/or submitted
- Sign into SeqLIMS.
- Go to the "Main" page and locate your order.
- Click on the "Edit samples etc." button to fill in information about your samples.
- Prepare your samples according to our Sample Requirements – we will return samples that are not properly prepared and/or submitted
Submit Your Sample(s) (Prepared Samples Required)
- Print a copy of your unique order number (not quote) and place that with your samples
- We have three ways to submit your samples
- Duke’s Campus:
- *email microbiomesr@duke.edu to coordinate drop off
MSRB3 - 3 Genome Ct., Durham
Mondays - Tuesdays, 2 p.m
**Any biological samples that need extraction services must be dropped off at Chesterfield and NOT MSRB3
- *email microbiomesr@duke.edu to coordinate drop off
- Direct Drop Off
Chesterfield Bldg - 701 W. Main St, Suite 320
Mondays - Thursdays, 10 a.m - 4 p.m.
Fridays 10 a.m. - 12 p.m.
- Ship
Attn: Sequencing Order [#xxxx]
GCB Sequencing and Genomic Technologies Shared Resource
Chesterfield Bldg.
Duke University
701 W. Main Street
Suite 320,
Durham, NC 27701
USA
- Duke’s Campus:
- We deliver your data in the form of demultiplexed FASTSQ files via an sftp server. We will send you login and password information to access the data via email. To share data with collaborators, you can simply forward the email containing the login and password information. Data is retained for thirty days. Please reach out to us about delivering demultiplexed 10x data.
- Most sequencing runs performed today are paired-end. This means you have a file from both ends of the insert - read 1 (R1) and read (R2). If your order was on more than one lane, each lane will produce a separate sequencing file also (L001-L004)
- Beyond delivery of FASTQ data, we provide Bioinformatics Services
- Mapping and Basic QC
- RNA-seq data processing
- Variant calling
- Peak calling
- For all publications that include data generated in the Sequencing and Genomic Technologies Core Facility, we kindly request that you acknowledge this support:
We thank the Duke University School of Medicine for the use of the Sequencing and Genomic Technologies Core Facility, which provided ______________ service.
All PacBio Revio results must include the NIH grant that was used to purchase the Revio:
We thank the School of Medicine for the use of the Sequencing and Genomics Core Facility for [insert service here]. Purchase of the PacBio Revio was funded by the NIH (1S10OD034222-01).