We are located on the third floor of the Chesterfield Building, 701 West Main Street in downtown Durham.
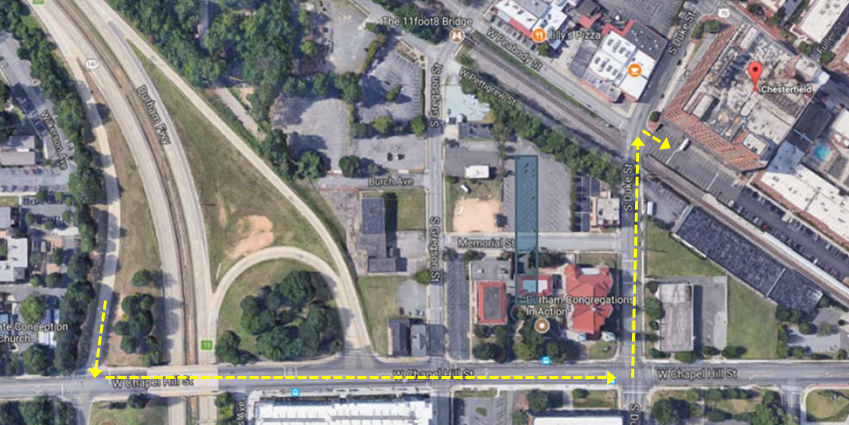
Directions to the Chesterfield Building
Duke Downtown Shuttle to the Chesterfield
- DukeCardID required for all passengers
- Operates from 7 A.M. - 6 P.M. Monday - Friday
- Track in real-time with Duke Transloc ("Shuttle: Downtown Pilot")
- Free WiFi is available via Carolina Livery
- Serviced by two 36-passenger shuttles operated by Carolina Livery
- Shuttles are ADA compatible
Parking at the Chesterfield
Duke visitors have a two-hour parking option, which is first come/first served access, to the visitor parking lot at the back of the building near the railroad tracks. We share this 2 hour parking with other tenant's visitors, and are not designated a certain proportion of parking for our guests at this site. Do not park beyond the fencing or cones; this is a construction area.
We have designated guest parking under the building and that can be reserved for up to six spaces for School of Medicine, and seven spots for School of Engineering per day. There are two calendars (below) that you can use to reserve parking spaces under the building. Please note if the current total of 13 spaces has already been met, then other accommodations will need to be made.
For larger events that might exceed the total number of parking or events lasting longer than two hours, we typically work with the building management to open up the 704 W Pettigrew St parking deck for more visitors on the day of the event.
1. Sample Submission Consultations
Prior to submitting any samples for new projects, please contact us. We utilize video conferencing solutions for most project discussions. We cannot accept samples without consultation and the electronic submission of the sample.
You can submit samples and check the status of submitted samples with PAMLIMS (see below).
If the samples are extensions of previous projects, please contact the scientist you previously worked with to discuss these samples.
Priority will be given to DCI members and time-sensitive projects, including preliminary data needed for timely grant submissions and additional experiments needed for manuscript resubmissions.
2. Electronic Sample Submission Process
Following a consultation with one of our staff members (see step 1 above), that staff member will send you a "PAMLIMS" lims system link to a) create an account and b) electronically submit your samples. There will be additional instructions within the PAMLIMS system.
** Note that existing projects and data currently in Express Repository has been archived. To retrieve data stored on Express, you will need to contact us and it will be re-mounted on DukeBox.
If you have any questions on PAMLIMS, please feel free to contact us or browse the "Q&A" section within PAMLIMS.
3. Procedures for Sample Delivery
- Submit your order into our system and prepare samples for delivery. You must contact the DPMCF at proteomics_metabolomics@duke.edu before submitting samples electronically. This will ensure our lab is ready to accept your samples.
- Samples can be delivered directly to the Chesterfield via prior arrangement and scheduling with your DPMCF contact. The address is 701 West Main St. and the entrance to the building is on the Main St. side. Free temporary parking is available in the rear of the building by taking a right immediately after the railroad tracks. Street parking is also available all around the site. Our scientists will meet you at the front door and collect your samples. Bring samples and a hard copy of the electronic submission(s).
- Use wet ice if samples need to be stored at 4oC.
- Use dry ice if samples need to be stored at -80oC.
- Ensure that the sample name on each tube matches the sample name on the hard copy of the order, and the order number of samples delivered matches the number of samples in your order.
- If shipping samples from outside of Duke University, please share shipping information with DPMCF so that they can be tracked from the receiving end as well. Please ship samples on dry ice if possible. The shipping address for samples sent by other delivery methods is below. Please ensure the samples have been submitted electronically before shipping samples.
Duke Proteomics and Metabolomics Shared Resource
c/o Ms. Laura Dubois
Center for Genomic and Computational Biology
Duke University School of Medicine
Suite 320
Chesterfield Building
701 W. Main Street
Durham, NC 27701
SDS-PAGE Gel-band Handling Recommendations
Although nearly any SDS-PAGE system can be utilized upstream of an LC-MS analysis, we recommend Invitrogen’s NuPAGE Bis-Tris mini-gel system. A good general purpose gel covering a large MW range (6-200 kDa) is the 4-12% gradient gel (Cat. No NP0321) using MES running buffer. These pre-cast gels provide excellent resolution, fantastic staining sensitivity (10 ng BSA using colloidal blue staining kit, Cat. No. LC6025), and are highly compatible with downstream LC-MS analysis. Additional product information.
We offer SDS-Page protein separation as a service. If we perform this step, we will share the image of the gel with you prior to cutting bands to ensure that the appropriate band(s) are excised and analyzed.
Pricing
Important reminders to avoid keratin contamination:
- Any gel manipulation prior to trypsin digestion should be done in a BSC or laminar flow hood.
- Wear nitrile (not latex) gloves.
- Wear a lab coat and make sure there is no gap between your coat sleeve and the gloves (lab tape works well).
- Ensure that gels are cut against a clean glass plate (methanol rinse) using a new, methanol-cleaned scalpel or razor blade. Please cut as close to the margins of the band as possible, to avoid background contamination from the non-stained region of the gel.