
Researchers at Duke University School of Medicine have unveiled a breakthrough genetic sensor that shines a light on critical changes in our genetic material.
The sensor, developed by a Duke team led by Kate Meyer, PhD, an assistant professor in the Department of Biochemistry, produces a fluorescent glow when it detects m6A, a small but powerful modification in RNA.
According to a Jan. 2 study in Nature Biotechnology, the first-of-its kind sensor could accelerate research as scientists explore the promise and power of RNA to combat diseases, among them COVID, cancer and heart disease.
“The idea to build an m6A sensor was first conceptualized back in 2019. However, as happens so often in research, turning an idea into a working system was harder than anticipated,” said Meyer, co-director of the Duke Center for RNA Biology who also holds secondary appointments in cell biology and neurobiology.
“After testing multiple different reporter mRNA strategies, one version of the system finally showed evidence that it worked,” she said.
Meyer is senior author of the study that describes the mechanisms of the genetically encoded m6A sensor (GEMS), which she designed and patented.
It targets m6A, an RNA modification that affects how genes work and how cells do their jobs. Whether m6A is present or not can influence an array of biological processes such as how we grow, how we learn and remember things, circadian rhythms, fertility, and immunity.
“We hope that this system will find widespread use among others in the research community and lead to insights into m6A biology."
- Kate Meyer, PhD
The game-changing aspect of the newly developed technology is in its ability to address a long-standing research limitation. There has not been an effective way to monitor changes in m6A within living cells.
All current methods require isolating RNA from cells and usually involve expensive steps like mass spectrometry and next-generation sequencing.
Enter GEMS: a straightforward and visually striking readout that lights up when RNA undergoes methylation, a chemical process that affects how the molecule functions in the body, and m6A is present or remaining dark when m6A is absent.
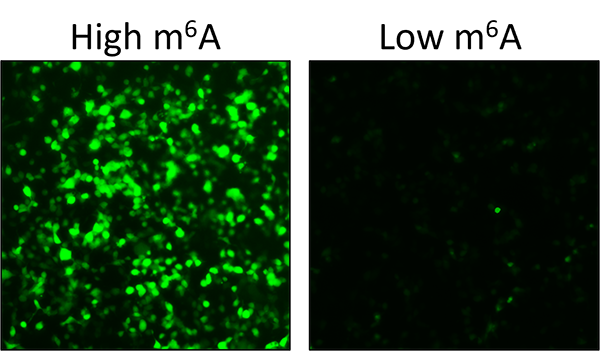
The technology provides new opportunities for scientists from studying how m6A behaves in different situations and illnesses such as COVID and cancer to quickly screening for drugs that could target the machinery behind m6A.
Imagine finding new treatments for diseases just by watching these tiny changes light up.
Accelerating RNA Exploration
GEMS provides researchers with a direct and accessible platform for real-time monitoring of m6A dynamics and may introduce a new era in studying RNA modifications.
“The development of the first genetically encoded m6A sensor is a big step forward because it can sense dynamic changes in RNA modifications within its native context and relies on a simple and reliable fluorescent tag to monitor levels of cellular m6A,” said the study’s first author Fadi Marayati, PhD, a postdoctoral associate in the Meyer Lab at Duke University School of Medicine.
The technology’s potential impact extends to cancer research, where scientists are exploring targeting the m6A RNA machinery to suppress cancer tumors, and investigating links between disturbances in m6A modification and cardiovascular disease and neurological disorders.
The Duke team is fine-tuning the system to increase its specificity and go beyond observation to use it as a tool to couple m6A levels with expression of other proteins of interest, such as tumor suppressors.
“We hope that this system will find widespread use among others in the research community and lead to insights into m6A biology,” Meyer said.
Additional Duke authors: Matthew Thompson, PhD, and Stacy M. Horner, PhD, in the Department of Integrative Immunobiology and Christopher L. Holley, MD, PhD, in the Department of Molecular Genetics and Microbiology.
Funding: The Rita Allen Foundation and the National Institutes of Health (R01MH118366, DP1DA046584 and RM1HG011563) funded the study.