
Building A Better Protein Trap
Life scientists love antibodies, not only because these little proteins help protect us all from pathogens, but because antibodies are also a very handy laboratory tool for identifying and marking proteins of interest in their research.
When you’re trying to find something very tiny, you need an itty bitty flag to mark it. That’s an antibody.
Like most life science researchers, Duke cell biology chair Scott Soderling has been reliant on custom antibodies, molecules made-to-order by hundreds of different supply labs that help scientists find and mark specific proteins in cell cultures and living organisms.
“But there's a problem,” he explains in the small conference room adjacent to his Nanaline Duke office. “Fifty percent of the antibodies on the market are junk. They're not specific. They might bind what you think they bind, but then they bind to other things you don't know about, or they don't even bind what you want to bind to at all.”
Worse than that, one batch of bespoke antibodies may not be the same as the last one. “Say you have a perfect antibody that binds exactly what you want and nothing else. And then you order the next lot and there’s a different preparation from a different animal, and you're back to square one. It doesn't work.”
“It’s thought that these bad antibodies lead to a large fraction of the irreproducible results,” Soderling says. “So it costs money, it costs time and it costs credibility. This is a huge problem for science, both academic and industry.” In part, the problem stems from the fact that custom antibody manufacturing techniques date to the 1970s, he says.
Soderling has founded a Duke spinout company he hopes will solve the reliability problem. CasTag BioSciences is based on a technology developed in his lab that marks proteins of interest in an entirely new way, using the genome-editing tool CRISPR.
One major thrust of Soderling’s research has been identifying proteins in the synapses of the brain, the tiny gaps between nerve cells where signals are transmitted and received. All that signaling is regulated by specific proteins. But identifying all of those proteins in the synapse and interpreting what they’re saying to the cell is a huge problem in a very tiny space. Antibodies are a key tool, but the work has been frustrating and slow, in part because of the difficulty of working with custom antibodies.
About three years ago, as news of the new gene-editing technology called CRISPR spread, Soderling and his team wanted to see if it might give them a better way to label and visualize the hundreds and even thousands of proteins they were detecting in the tiny synapse between neurons.
“We had this idea that CRISPR could be a really amazing tool to address the pressing problem of trying to identify and label these hundreds of proteins,” Soderling says. “What we developed was a new modular method for basically taking the labeling problem and flipping it on its head.”
They’re using CRISPR to edit short sequences into a gene so that every protein it produces carries a tag they have created that is detected by a known, reliable and well-characterized antibody, rather than a shot-in-the-dark custom antibody.
“These antibodies recognize a small segment of amino acid sequences,” Soderling explains. “So we just take the DNA encoding those amino acids -- the handle -- and we plop that handle right into the gene in vivo, or in the cell,” Soderling says.
After the proof-of-concept experiments produced beautiful protein labeling in the mouse brain, Soderling looked at the images and said, “Okay it’s huge.”
Indeed, they dubbed their new system HiUGE (homology-independent universal genome engineering), and it might just be huge.
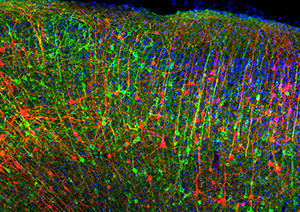
“Homology-independent Universal Genome
Engineering”, or HiUGE, uses adeno-
associated viruses to deliver multiple “plug
and play” gene sequences to a variety of
cells in a lab dish or a living organism. (The
colored neurons in this image are in a mouse
brain.)
They’ve taken to calling it plug and play biology, because with just a few of their tags, they can address hundreds of unknown proteins, and they can even put multiple tags into a gene at the same time. Soderling says the system is modular and easy to use, which will enable semi-automated, high-throughput approaches to labeling proteins.
By way of analogy, think of a delivery truck driver going slowly down the block after dark in a downpour looking for house number 2345. What Soderling and his team have done is put a bright sign on every house numbered 2345 that says “Hey UPS! Over here!”
The HiUGE system is delivered to living cells, either in a dish or in an organism, by a pair of adeno-associated viruses working as a team. One virus carries guide RNA which will mark the spot at which CRISPR should cut the DNA and insert a new piece of code. The second adeno-associated virus carries ‘the payload,’ a tag or tags they’ve devised that will now be built into every protein that gene subsequently produces.
The vectors, including a synthetic guide RNA and HiUGE tags, are agnostic, or ‘homology-independent,’ as the name implies. They don’t care what gene is around them. “We designed this guide RNA so that it specifically doesn't recognize anything in the mouse, human, monkey, cat or donkey genomes,” Soderling says.
It’s a clever way to explore the unknown.
Not only does this approach advance their own work, Soderling began to realize that a fast, flexible, more accurate way to tag proteins might also be a business opportunity. With a little research, he figured out that custom antibodies are a $2.4 billion market – again, with products that only work as advertised half the time.
He reached out to Duke’s Office of Licensing and Ventures (OLV) to begin the patenting process and to get some advice on starting a company. “Then I had to find a way to run the business, because I already have a great day job.” In fact, he had also just been named chair of cell biology at about the same time.
At OLV’s recommendation, Soderling visited Biolabs North Carolina, a shared workspace in the Chesterfield Building in downtown Durham which leases individual wet-lab benches on a month-to-month basis and provides all the basic equipment a startup would need, including refrigeration, gene-copying PCR machines, centrifuges, etc. He pitched his idea to Biolabs and had a look around.
The next day, BioLabs NC president Ed Field called Soderling and asked if he’d like some help running the business. Field, a startup veteran, is now the CEO of CasTag. The firm has raised enough money with a loan from the North Carolina Biotechnology Center to hire a recent Fuqua Business School graduate as the business development lead and a former postdoc for Soderling to run the lab part-time while he looks for a job in industry.
“We've got a website. We've got orders. We've got customers. It's up and running,” Soderling says, with a measure of wonder in his voice. His conference talks about HiUGE and a July 1, 2019 paper in Neuron attracted some attention. Then the paper was republished as one of the journal’s “best of 2018-2019”, drawing still more notice.
And now they also have ideas for new products. “I'm hoping that this will expand and become even bigger than just tagging proteins,” Soderling says.
“You know, North Carolina was a manufacturing state back in the day,” says Soderling, a soft-spoken native Tennessean. “I would love to wake up some day and drive into downtown Durham and see one of the former manufacturing warehouses humming away with people making these reagents to ship out around the world. That's the dream.”
Karl Leif Bates is the Director of Research Communications at Duke University.
Photos of Scott Soderling and his lab were taken by freelance photographer Les Todd.
