Tuberculosis (TB) infects more than 10 million people each year worldwide, causing 1.25 million deaths. In 2025, the disease made a troubling comeback in the United States, including a rise in cases in North Carolina for the first time in decades.
In some people with the disease, clusters of immune cells called granulomas form inside the lungs. These microscopic “cities” can harbor the bacterium that causes TB, allowing it to persist and resist antibiotics.
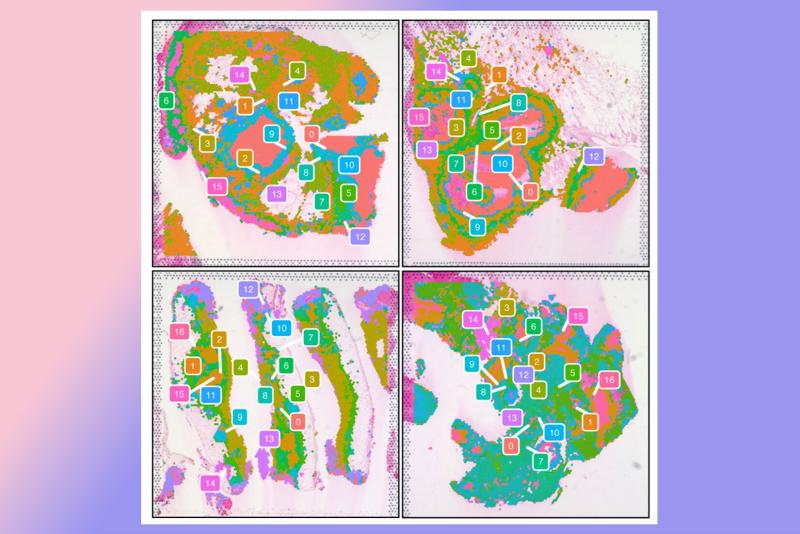
Now researchers at Duke University School of Medicine have used advanced genetic sequencing tools to show the identity of all the “residents” of these cellular cities, pinpointing their exact locations and how they interact.
The map revealed that a particular protein, osteopontin, plays a crucial role in building granulomas. And the study offers a rich dataset that could pave the way for new treatments.
The results were published online in the journal mBio.

“The osteopontin finding is an example of the power of these data to reveal how granulomas function,” said David Tobin, PhD, professor of molecular genetics and microbiology and senior author of the study. “And it wouldn’t have been possible without a team of Duke collaborators.”
Granulomas can be “both a prison and a penthouse,” for the TB bacterium, said first author Charlie Pyle, PhD, PharmD, a senior research associate. They trap the bacteria and stop it from spreading, but they can also block immune cells and antibiotics from reaching and killing the infection.
So, when Pyle learned that Duke infectious disease specialist Jason E. Stout, MD, MS, and pathologist Jadee Neff, PhD, could provide archived tissue samples from biopsies of patients who turned out to have TB, he saw an opportunity to map the immune cells inside granulomas in unprecedented detail.
One of the tools Pyle used: spatial transcriptomic profiling, a technique that shows how genes are expressed across an entire piece of tissue, not just in isolated cells.
“We mixed together the spatial and the single-cell data and found out where in space particular cells are,” he said.
From this map, a pattern emerged. Cells called macrophages predominated, particularly ones that express the protein osteopontin.
The team confirmed the crucial role for osteopontin in a zebrafish model of TB. Infected fish genetically engineered to lack the protein struggled to form granulomas and had shorter lifespans than those with it.

“It’s a massive amount of data: almost every gene that the body makes and how it’s expressed in these granulomas,” Pyle said. “A few years ago, this would have been a dream for me, to look at this lesion and find out what exact molecules are in which cells, and where.”
He and other researchers can now use the information to explore how to resolve granulomas without the risk of harming patients, Pyle said.
Other authors of the study: Liuyang Wang, Rebecca W. Beerman, Vaibhav Jain, Henry K.E. Ohman, Brandon A. Thompson, Karen R. Abramson, Dennis C. Ko, Simon G. Gregory, Clare M. Smith, Rebecca J. Richardson.
Funding: That National Institutes of Health, the British Heart Foundation, and the Duke University Center for AIDS Research.