Structural insights about a deadly bacterium’s tools point to ways to block it
The bacterium that causes the tick-borne disease tularemia is a lean, mean infecting machine. It carries a relatively small genome, and a unique set of infectious tools, including a collection of chromosomal genes called ‘the pathogenicity island.’
A team of researchers from Duke University, Harvard, University of Rhode Island and the National Institutes of Health has now unpacked the bacterium’s toolbox and built an understanding of the shapes and interactions of all its parts, using an imaging technique called cryo-electron microscopy.
Their insights, which appear Nov. 19 in Molecular Cell, point to a way in which the bacterium’s unique infectious machinery might be blocked.
The bacterium Francisella tularensis can infect more than 200 kinds of animals including humans, dogs, cats, fish and rodents. As few as ten bacteria, usually from an animal or insect bite, are sufficient to be an infectious dose. Tularemia attacks skin, eyes, lymph nodes and lungs, sometimes leading to pneumonia, meningitis, inflammation around the heart and bone infections. Fortunately, it is uncommon in humans, but it can be deadly and make people very sick.
In fact, since the 1920s, Russia, the U.S. and other countries have studied it as a potential bioweapon.
The bacterium is capable of infecting many different cell types, but its sweet spot is the macrophage, an immune system cell that is a first-responder to invading bacteria.
When an alien bacterium is detected, a macrophage will envelope the bug and capture it within a bubble of its cell membran called a phagosome, where it will be chewed up and rendered impotent. That’s usually the end of the story.
But Francisella, sensing it has been captured, turns on a specialized set of its own genes, the pathogenicity island, and begins making the tools it will need to escape from the phagosome and enter the macrophage’s fluid center, where it can replicate in peace.
“It’s kind of a professional pathogenic bacterium,” said Maria Schumacher, a distinguished professor of biochemistry in the Duke School of Medicine, who is co-corresponding author of the study. One of the key parts of its infectious machinery is a protein called MglA (macrophage growth locus protein A), which has been found nowhere else. “This bug has evolved these special proteins,” Schumacher said. “It’s also very sturdy; it can survive in dead carcasses and soil too.”
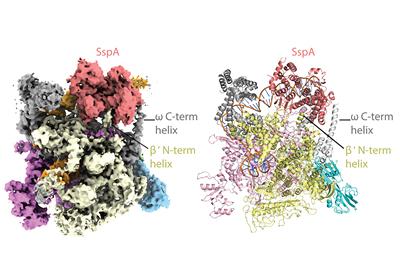
The researchers used state-of-the-art cryo-EM microscopes at Duke and Harvard to see the shapes and interactions of all the bacterium’s infectious machinery and how they worked together.
The researchers were able to determine the shapes and interactions of all the players in the system, from the small molecule signal to the large multi-protein complex, a “virulence specialized RNA polymerase,” that activates the genes on the pathogenicity island.
“It’s a novel system and that’s why we’re so excited about it, Schumacher said. “It’s not like any other that we’ve found in any other bacterium or eukaryote. It’s altogether very unique and interesting.”
“Even if you know all these components, how they work together is not clear,” Schumacher said. “We could actually look at it and see okay, how is this working?” The key structural analyses on the large molecular complexes were performed by Brady Travis, a Biochemistry graduate student in the Schumacher laboratory and first author of the study, who mastered cryo-EM to allow the visualizations of these complexes.
Schumacher and co-corresponding author Richard Brennan, chair of biochemistry at Duke, are now pursuing a grant to screen a collection of small molecules that might bind to MglA and prevent it from allowing the assembly of this specialized complex for virulence.
“We found a nice pocket for binding potential inhibitors of MglA function,” Brennan said. “If you can prevent it from binding to its partner, you won’t get the pathogenicity island transcribed, which is a good thing.”
This research was supported by the U.S. National Institutes of Health, Department of Energy, and National Science Foundation. (R35GM130290, R21AI146641, AI081693, AI145954, F31AI150138, HD055148-08, U24GM129547,DE-AC02-05CH11231, ECCS-1542015)
CITATION: "Structural Basis for Virulence Activation of Francisella tularensis," Brady A. Travis, Kathryn M. Ramsey, Samantha M. Prezioso, Thomas Tallo, Jamie M. Wandzilak, Allen Hsu, Mario Borgnia, Alberto Bartesaghi, Simon L. Dove, Richard G. Brennan, Maria A. Schumacher. Molecular Cell, Nov. 19, 2020. DOI: 10.1016/j.molcel.2020.10.035
This article was first published in Duke Today