Antibodies are like dogs. Some, like show dogs, have the perfect construction and temperament, but instead of winning Best in Show, the top antibodies more effectively fight off viruses and other infections. Not all antibodies are “show quality,” though. Some may not be as potent or broad enough to win, but these “pet quality” antibodies are still great companions that can help fight disease, albeit less efficaciously.

Bruce Donald, PhD, James B. Duke Distinguished Professor of Computer Science, and team are using computation strategies to figure out ways to turn those “pet quality” antibodies into “show quality” antibodies to better fight against HIV. Results are published in Cell Reports.
In a close, years-long collaboration with scientists led by Peter Kwong, PhD, at the National Institutes of Health (NIH) Vaccine Research Center (VRC), the team had previously discovered some promising antibodies to fight against HIV, but, in order to be truly effective, they needed to up their game.
“Better, more potent and broader antibodies,” Donald said, “can be directed at different sites of the HIV virus to better neutralize it.”
Like the virus that causes COVID, HIV has spike proteins. Different antibodies attack the spike proteins in different places, so having multiple antibodies attacking different parts of the spike proteins helps kill the virus more efficiently and with less risk of developing resistance.
When the first promising antibodies to fight HIV were discovered, it didn’t take long to learn that they fought off a lot of the strains of HIV, but not the most difficult ones. “At clinical concentrations,” Donald said, “they would neutralize about 75% of the viruses.” That means if you treated HIV with that antibody only, the other 25% of the viruses would evade the treatment and get stronger. “You would be selecting for resistance,” Donald said.
Having antibodies that are both potent and broad helps mitigate issues with resistance, but up until now, ensuring antibodies are broadly neutralizing has been tricky. Donald’s team figured out how to improve the breadth of HIV antibodies. “This is perhaps the first time,” Donald said, “that there has been a systematic attempt to improve mechanisms of antibody breadth based on geometric and physical computation.”
The Donald lab’s software, OSPREY, is an open-source computational software that can design protein structures. By employing OSPREY, the team improved the characteristics of naturally occurring broadly neutralizing antibodies against HIV. “Using the software ‘out of the box,’” Donald said, “we were able to run the program, predict antibodies to test, test the antibodies in a biological assay, and prove that they were broader and more potent on a large panel of viruses.”
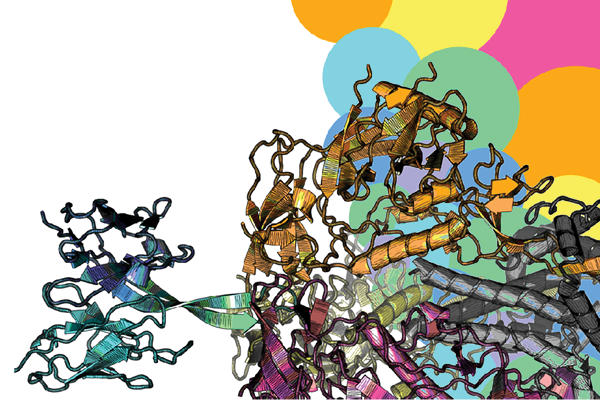
Basically, by modifying naturally-occurring “pet quality” antibodies that were already known, the researchers figured out how to turn them into stronger, broader “show quality” antibodies. Then, using cryogenic electron microscopy (cryo-EM), they solved the structures of three of the best designed antibodies in high resolution to show the mechanisms of improved potency and breadth and how the antibodies bind to the virus.
“It’s amazing that you can let OSPREY think about it and propose changes to the antibodies that will make them broader and more potent,” Donald said, “and then take those results and test them in the lab to confirm with high resolution structural information.”
They tested their designer antibodies on a panel with 208 strains of HIV and found that overall, these enhanced antibodies are four times more potent, and with some strains, they are 100 to 1,000 times more potent with significant improvements in breadth as well.
“So, if you discover antibodies that are promising but not quite as broad or potent as the best ones,” Donald said, “then using computation, we can redesign the antibodies and bring them up to the very top of that standard.”
Now that the antibodies have been created, tested for both breadth and potency, further preclinical work can be done to continue to test efficacy, before animal models and eventually clinical trials for new therapeutics to treat patients with HIV.
“We now have overwhelming data to support that our designs work,” Donald said. “This shows the power of computation!”
Image caption: An antibody designed by Donald and team binds to HIV spike protein.