Striped gene expression pattern provides clues for neurodegenerative movement disorders
Spinocerebellar ataxias are a family of neurodegenerative disorders affecting muscle coordination and control. Because the brain is such a complex organ, these disorders have been poorly understood.
Duke researchers are using single cell RNA sequencing technologies to better understand how these diseases form, which may lead to better treatment options for patients. Recent results were published in Science Translational Medicine.
Spinocerebellar ataxia 7 (SCA7) is one of nine disorders in this family of polyglutamine disorders. They are genetically inherited and are caused by repeated CAG building blocks of DNA, which codes for the amino acid glutamine. If the repeated CAG sequence crosses a threshold of 36 or more repeats in the ataxin 7 gene, patients begin to develop symptoms associated with these disorders, such as a progressive loss of balance and coordination, slurred speech, slowed movements, stiffness, tremors, and an inability to decipher how far away objects are.
The severity of the disease depends on the number of repeated CAGs in the DNA. Because this is an inherited disease, each generation can compound the number of repeats, making the mutation worse. “We see patients who have lived essentially normal lives until their 60s, and then they start experiencing symptoms,” said Luke Bartelt, lead author and postdoctoral scholar in the lab of Craig Lowe, PhD, assistant professor of molecular genetics and microbiology. “But generally, their kids and grandkids will get the mutation worse with each generation.” This means that while a grandparent may not experience symptoms until late in life, their child might start seeing symptoms in their 40s.
Using a mouse model of SCA7, the team conducted single-cell RNA sequencing in the cerebellum. “We were able to quantify all of the genes, the gene regulation or RNA produced from all of these genes within each cell type,” said Bartelt.
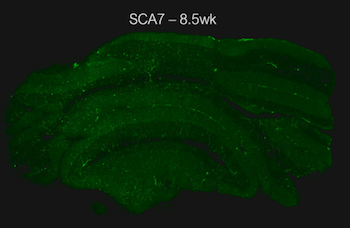
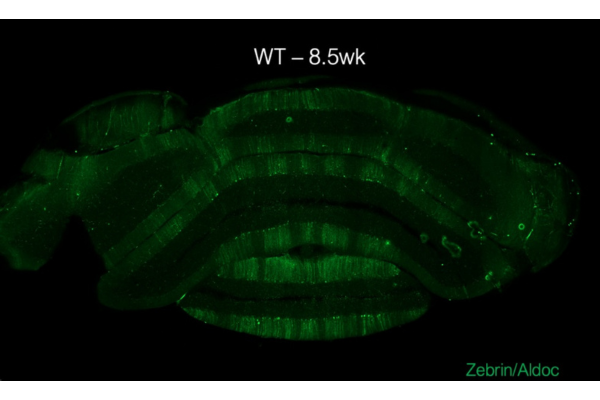
They observed a critical disruption in gene expression within Purkinje cells, large neurons in the cerebellum essential for coordinated movement. “These neurons are normally organized in a characteristic striped gene expression pattern, partly due to the zebrin-II gene Aldoc, whose expression forms visible stripes across the cerebellum,” said Bartelt. “Our research showed that SCA7 disrupts these stripes, causing them to disappear after symptom onset.”
Since Purkinje cells are rare, the team needed a way to enrich the capture of these cells. “Even with single cell technologies,” Lowe said, “we can only sequence so many cells, and when a cell type is really rare, it’s hard to capture very many of that cell.” So Bartelt developed a way to capture those cells more efficiently and get a more even distribution of cell types. This technique enables a more refined analysis of cerebellar cells, which could allow for broader applications in studies of both disease and normal brain function.
The striped gene expression pattern observed in Purkinje cells was not only disrupted in SCA7 but also in other polyglutamine spinocerebellar ataxias such as SCA1, SCA2, and SCA3. This suggests that therapeutic strategies could potentially target multiple forms of ataxia and pave the way for treatments that address a range of polyglutamine diseases, possibly easing the therapeutic development process for this challenging group of conditions
Currently, treatments for this family of disorders are limited to symptom management. This research may lead to the development of gene therapies that selectively address dysfunctional cells, such as Purkinje cells in SCA7. This approach might enhance therapeutic precision and offer improved outcomes compared to generalized treatments.
“Our research shows that we can’t treat all cells the same. What might help one neuron might hurt a different type of neuron,” Bartelt said. “This is important information to be able to develop cell type-specific gene therapies and figure out a way to correct the regulation of Purkinje cell subtypes.”
In addition to Bartelt and Lowe, study authors include Albert La Spada, Court Hull, Harry Orr, Stefan Pulst, Daniel Scoles, Hayley McLoughlin, Sabrina Jarrah, Lisa Duvick, Fabiana Longo, Grazyna Adamek, and Pawel Switonski.